Proteins and Proteomics
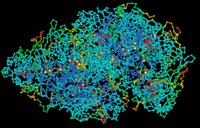 Proteins are complex, macromolecules comprised of amino acids linked by peptide bonds into long chains. The sequence (primary structure) and properties of constituent amino acids generate the 3D conformational structure (tertiary and quaternary structure) that is vital to the biological function of proteins. (click to enlarge image)
Proteins are complex, macromolecules comprised of amino acids linked by peptide bonds into long chains. The sequence (primary structure) and properties of constituent amino acids generate the 3D conformational structure (tertiary and quaternary structure) that is vital to the biological function of proteins. (click to enlarge image)Proteins are essential to the structure and biological viability of all living cells and viruses. The cellular proteome is the total cellular protein under a particular set of conditions, while the complete proteome is the sum of all potential proteomes of a cell. Proteomics has become the subject of much research in cell and molecular biology.
Proteins play a number of vital roles as:
a. Enzymes or subunits of enzymes – catalyzing cellular reactions.
b. Structural or mechanical roles – structural components of tissues, components of the cytoskeleton, centrioles, cilia and flagella, microtubules, molecular motors.
c. Intracellular and intercellular signaling functions – ion channels, receptors, membrane pumps.
d. Regulatory proteins or adaptor proteins in genetic transcription, RNA processing, spliceosomes.
e. Products of immune and inflammatory responses that aid in targetting of foreign substances and organisms.
f. Storage and transport of various ligands.
g. The source of essential amino acids.
Almost all natural proteins are encoded by DNA, which is transcribed and processed to yield mRNA, which then serves as a template for translation by ribosomes on the rough endoplasmic reticulum.
Protein roles:
● cytoskeletal protein, enzymes, ion channels, latent gene regulatory proteins, receptor proteins, 'signaling' enzymes, signaling proteins, transcription factors
● activator,
● adaptor protein, amplifier protein, anchoring protein, bifurcation protein, coincidence detector protein, effector protein, messenger protein, modulator protein, relay protein, scaffold protein, transducer protein
Specific proteins/types : § adaptor protein : cAMP receptor binding protein § CARD domains : cofactor § collagen : core histones H2A, H2B, H3, and H4 : CRE-binding protein CREB § C-reactive protein § CRP § domains : elongation factor EF § granulysin : helicases : Helicase II : heterochromatin : histone : HP1 § immunoglobulin isotypes : inducible transcription factors : LexA repressor : mCAT2 receptor : motor proteins § NF-κB : nucleosome : PcG proteins : PCNA § (pentraxins, CRP) § PH family : Polycomb group : proteome : RecA : regulatory proteins : repressor proteins : ribosomes : RPA : serine rich (SR) splicing factors : silencers : Ski7p : small nuclear ribonucleoproteins (snRNPs) : spliceosome : SR (serine rich) splicing factors : trans-acting factors : trithorax group (trxG) : UPF1 UPF2 : upstream transcription factors :
Enzymes ♦ Enzymes : ♦ activation-induced (cytidine) deaminase ♦ adenylate cyclases ♦ AID ♦ Akt : AP endonuclease (Ape1) ♦ ATPases ♦ bonds ♦ cAMP-dependent protein kinase ♦ cyclin-dependent kinases : DNA glycosylase : DNA Ligase I : DNA polymerases : DNA polymerase I : DNA polymerase beta : DNase IV ♦ energetics : exonuclease 1 : exosome : Fen1 : Flap Endonuclease FEN-1 ♦ Fyn : general transcription factors ♦ GTPases : hOGG1 : hOGG1 oxoG repair : LigIII : MAP kinase ♦ MAPKs ♦ MEK ♦ MPF : Msh2-Msh3 ♦ mTOR : MutS, MutL, and MutH : 8-oxoguanine glycosylase : oxoG repair hOGG1 : PCNA ♦ PDK1 ♦ PTEN ♦ PDK1 ♦ phosphatases ♦ phospholipases ♦ phosphodiesterases ♦ phosphorylases ♦ PIKK ♦ PI3K ♦ PKA ♦ PKB ♦ PKCs ♦ protein kinases ♦ reaction energetics ♦ receptor tyrosine kinases : RNA polymerase: Replication factor C : reverse transcriptase : ribozymes : RNA polymerase II ♦ serine/threonine kinases : spliceosomal-mediated RNA trans-splicing SMaRT : trans-splicing ribozymes ♦ UNG2 ♦ uracil-DNA glycosylase : UvrD : XRCC1 :
[] Rediscovering Biology - Animations and Images []
Alphabetic links to Glossary items:◦ A ◦ B ◦ C ◦ D ◦ E ◦ F ◦ G ◦ H ◦ I ◦ J ◦ K ◦ L ◦ M ◦ N ◦ O ◦ P ◦ Q ◦ R ◦ S ◦ T ◦ U ◦ V ◦ W ◦ X ◦ Y ◦ Z ◦ o-o animation index o-o diagram index o-o image index o-o micrograph index o-o table index o-o sem/tem index o-o video index .
tags [Proteins] [Proteomics]
Labels: cytoskeleton, enzyme, immune, proteome, regulatory, signaling
| 0 Guide-Glossary